Construction of a gene regulatory network (GRN) for the lens
We have taken two approaches to construct a comprehensive gene regulatory network for the lens. In our first approach, we aim to reconstruct a spatiotemporal GRN in mouse lens development based on the published literature. Our approach is based on one that has recently been utilized to construct a highly effective GRN for the tooth (O’Connell et al. Sci Signal. 2012 Jan 10;5(206):ra4. doi: 10.1126/scisignal.2002414, http://www.ncbi.nlm.nih.gov/pubmed/22234613). We are currently modifying this approach to the lens, by which we can reconstruct a useful GRN by effectively proposing data in the published literature. This will be a critical first step to generate a useful GRN. The advantage of this approach is that EVERY edge in the network can be traced to at least one piece of experimental evidence.
Data from the published literature on the lens has been collected as follows:
- Across 4 tissue compartments
- Epithelium (Epi), equatorial zone (EZ), fiber cells (FC), and optical vesicle (OV)
- Across 3 dev. Stages
- Specification / induction (S/I)
- Primary fiber cells differentiation (PFCD)
- Secondary fiber cells differentiation (SFCD)
- 572 separate evidences on gene regulation in the lens based on 20+ years of published literature.
Below, we have represented 4 tissue compartments (the lens epithelium in light green, the lens equatorial zone in dark green, the lens fiber cell in orange and the optic vesicle in light blue), across 3 major developmental stages (lens induction, primary fiber cell differentiation, and secondary fiber cell differentiation).
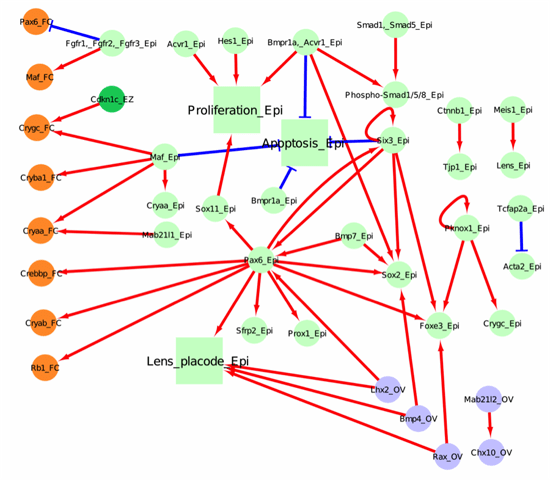
Figure 1. Network for lens induction
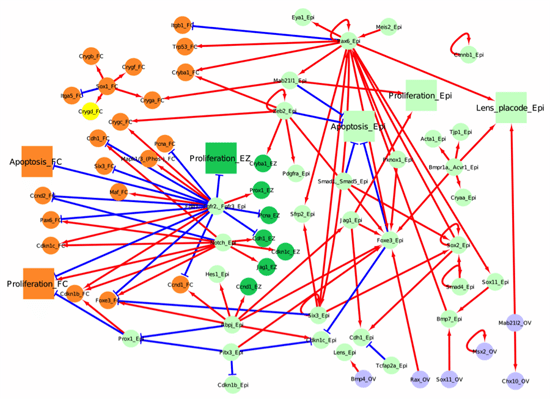
Figure 2. Network for primary fiber cell differentiation
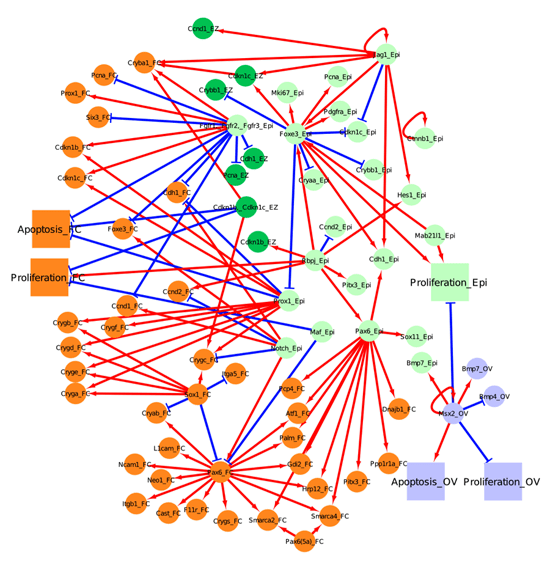
Figure 3. Network for secondary fiber cell differentiation
Although these are currently “static” representation of the network, they will subsequently be replaced by user-friendly interactive versions similar to that generated for ToothCODE (O’Connell et al. Sci Signal. 2012 Jan 10;5(206):ra4. doi: 10.1126/scisignal.2002414, http://www.ncbi.nlm.nih.gov/pubmed/22234613).
Expanding the lens GRN
In addition to assembling a literature-based lens GRN, we have undertaken an integrated approach comprising computational and biological components to extend the currently understood GRN.
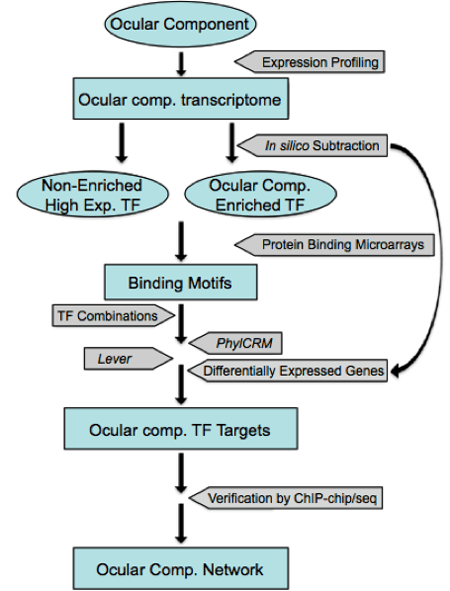
Figure 4. Strategy to build a comprehensive gene regulatory network in the lens (modified from Lachke and Maas 2010, http://www.ncbi.nlm.nih.gov/pubmed/20836031)
We are using computational prediction and chromatin immunoprecipitation (ChIP) coupled with whole genome expression profiling analysis to identify new nodes and to build a comprehensive GRN for the lens. Our strategy is outlined in Fig. 4. 1) We will use iSyTE to identify transcription factors (TFs) that show a dynamic expression pattern in the developing lens. The expression of novel genes of interest will be confirmed using in situ hybridization and IHC analysis. 2) The candidate TFs that survive these filters will then be used in a directed cis-regulatory module (CRM) analysis. Binding motifs for individual lens TFs or TF families will first be extracted from available databases including JASPAR, TRANSFAC, and UniProbe (http://thebrain.bwh.harvard.edu/uniprobe/). We will then use the Lever and PhylCRM algorithms to determine which combinations of TFs are likely to regulate differentially expressed lens-specific genes, and which DNA regions are their target enhancers (or CRMs) (Warner et al. 2008). 3) ChIP-chip experiments in the mouse lens epithelial cell line (21EM15) may also be used to identify downstream targets of these TFs. 4) The potential CRMs and their regulatory TFs identified will be validated in transient transgenic mouse experiments and used to construct the GRN.
