iSyTE – integrated Systems Tool for Eye gene discovery
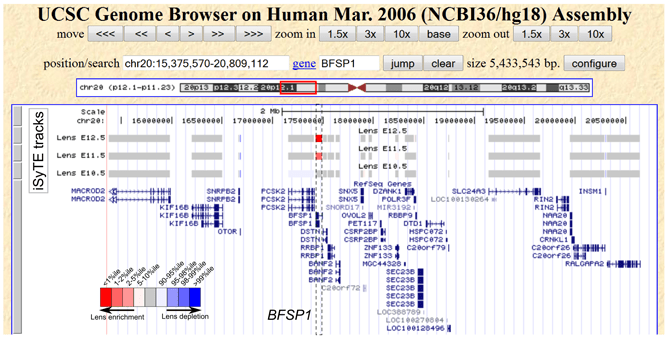
Figure 1. UCSC genome browser view of an genomic interval containing BFSP1, a gene associated with non-syndromic cataract
Using iSyTE tracks, we can easily identify that BFSP1 has the highest lens specific expression among all the genes in this mapped genomic interval.
The paper on iSyTE (Lachke et al. 2012 IOVS in press, https://www.ncbi.nlm.nih.gov/pubmed/22323457) is now published in Investigative Ophthalmology and Visual Science, click here for a PDF
To open a specific genome assembly on the UCSC Genome Browser with iSyTE tracks, please click on the following links:
Introduction
In the past century, our understanding of eye development and disease has come a long way and the genetic circuitry operational in the early stages of the ocular lens formation, for example, can now be derived in considerable detail. This circuitry presently comprises around 50 genes, most of which were identified only within the past 20 years. Identification of majority of these candidate genes has relied on insights gained from eye development research in Drosophila or on approaches involving the characterization of mutations associated with mouse and human ocular disease.
The identification of genetic mutation in human patients is traditionally based on linkage analysis and sequencing of candidate genomic regions in patient tissue or from relevant animal model. This is typically a time-consuming, labor-intensive process and prioritization of candidate genes within an interval is often tricky. With the advent of next generation sequencing, it is possible to rapidly identify a large number of genetic mutations in a sample, but it is still unclear how one can effectively identify the disease-associated mutation within this wealth of data. Additional biological knowledge is necessary to separate disease-associated genetic mutations from those that are not relevant to the phenotype of interest. Since many human genetic diseases are caused by aberrant developmental processes, we hypothesized that using knowledge of gene expression patterns during embryonic development in a mammalian animal model will yield key information for identifying disease-associated genes in human.
We present a simple yet effective experimental and computational strategy to prioritize candidate disease associated genes based on microarray gene expression profiling on embryonic tissues. To make this tool available to the community, we have developed a web-based public resource termed iSyTE (integrated Systems Tool for Eye gene discovery) that allows efficient identification of genes associated with congenital cataract. The present version of iSyTE integrates biological and computational components on the ocular lens, and allows the rapid identification of genes that are associated with lens development and disease, namely congenital cataract.
What is iSyTE based on?
iSyTE utilizes microarray gene expression profiles of the mouse embryonic lens as it transitions from the stage of placode invagination to that of vesicle formation. We identified differentially regulated genes by comparing lens microarray profiles to those representing whole embryonic body (WB) without ocular tissue. These were then utilized to generate a ranked list of lens-genes enrichment, which can be viewed as iSyTE tracks in the UCSC Genome browser to aid identification of genes with lens function.
What is the significance of the WB reference dataset?
We hypothesized that the WB may represent an ideal average gene expression profile for a mixture of tissues, and comparison of tissue-specific profiles against the WB control may therefore facilitate identification of tissue-specific gene expression. We have shown that the resulting in silico subtracted mouse lens database is an elegant tool to identify lens-enriched genes that play key roles in lens biology, and for identification and prioritization of potential candidate genes harboring mutations at mapped human cataract loci. This is consistent with the idea that selective gene expression in a tissue may be reflective of a function in the development or function of the tissue.
How well does iSyTE work for disease gene discovery?
When iSyTE was tested on previously mapped intervals of 24 genes associated with isolated or non-syndromic congenital cataract in human, it identified the known cataract associated genes in 88% of the cases by ranking it within the top 2 genes among all candidate genes in the locus. Gene set analysis demonstrates that this strategy can robustly remove non-specific, yet highly expressed, housekeeping genes in the lens tissue microarray profiles, which allows non-highly expressed lens-disease associated genes to be readily identified. In situ hybridization analysis has confirmed high lens expression of several novel iSyTE-identified candidate genes.
Using iSyTE for prioritization of genes within a mapped interval for cataract
iSyTE tracks (Human hg19, Human hg18, Mouse mm9, Mouse mm8) allows the visualization of genes in context of their enrichment of expression in the lens. After opening the browser for the human genome assembly window, the user can type in the interval of interest and the entire genome track is loaded. This representation allows immediate visual detection of the best candidates in a given genomic interval, and allows one to zoom in or out to visualize the presence of promising candidates within a particular region or proximal to it. In addition, visualization of the tracks representing three embryonic stages in one frame allows appreciation of the dynamic pattern of gene expression in lens development.
Extending the iSyTE approach to other tissues
We also investigated whether the iSyTE approach can be used to prioritize candidate genes in other organ. For this purpose, we generated and analyzed microarray profiles of embryonic tooth germs and matched WB controls. Gene set analysis reveals that in fact this approach can also effectively identify genes that known to be associated with tooth development and various human craniofacial defects. We created custom UCSC genome browser tracks, and they are available in the following genome assemblies: Human hg19, Human hg18, Mouse mm9,Mouse mm8.
Future: iSyTE for other ocular components and tissues.
Future versions of iSyTE will include data on other ocular components and will expand the datasets on the lens. We believe that in combination with other genome-wide datasets, network of validated gene regulatory relationships, and effective use of bioinformatics algorithms, iSyTE can be utilized to aid understanding of ocular development and pathogenesis, as well as for the identification of eye disease associated genes. We are currently pursuing several different approaches for construction of a gene regulatory network (GRN) for the lens. Please click here for the lens GRN.
Contact:
Salil Lachke, Ph.D.
Assistant Professor
Department of Biological Sciences
Center for Bioinformatics and Computational Biology
University of Delaware
Newark, DE 19716
E-mail: salil@udel.edu
